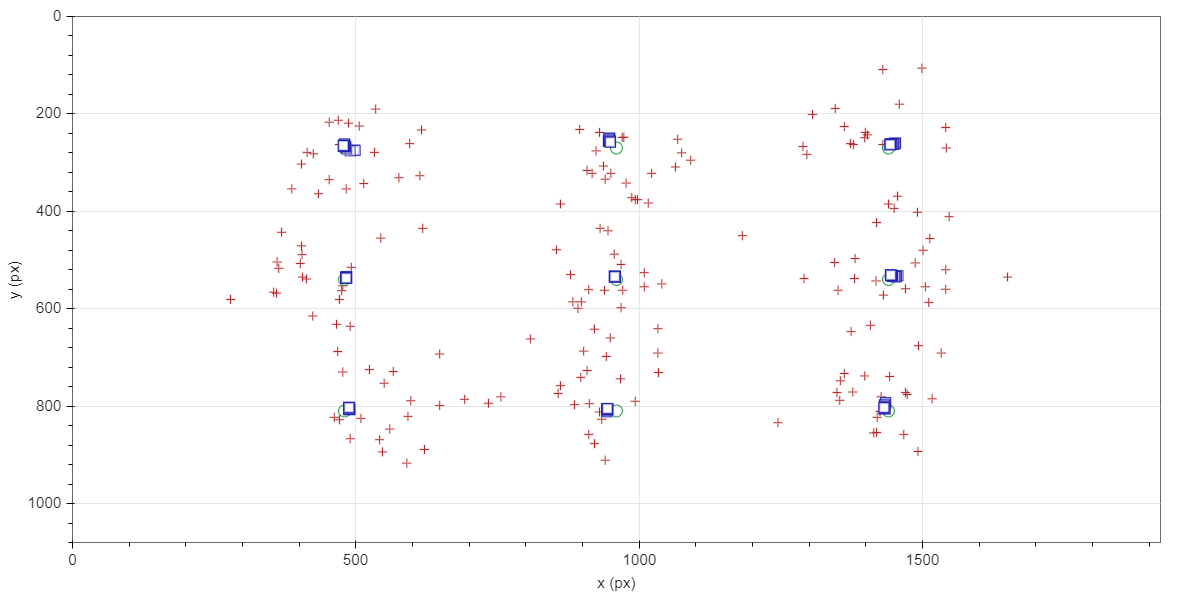
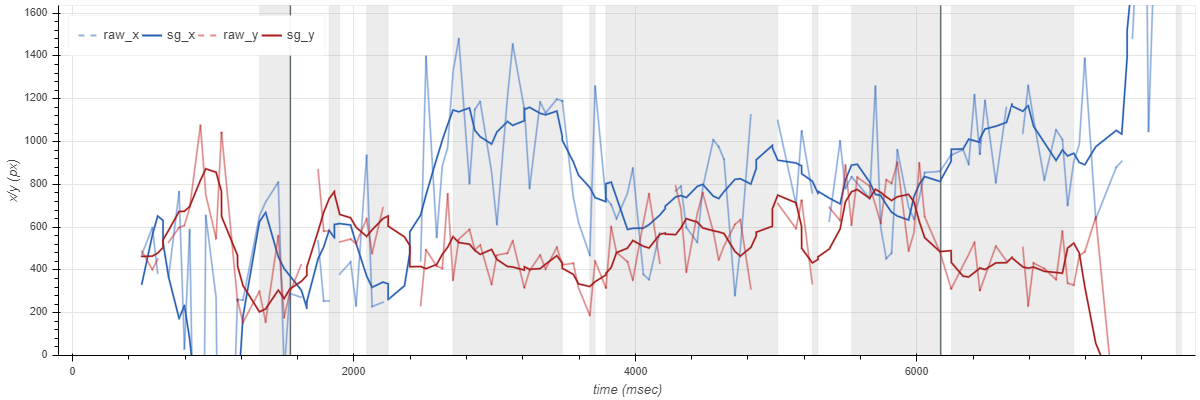
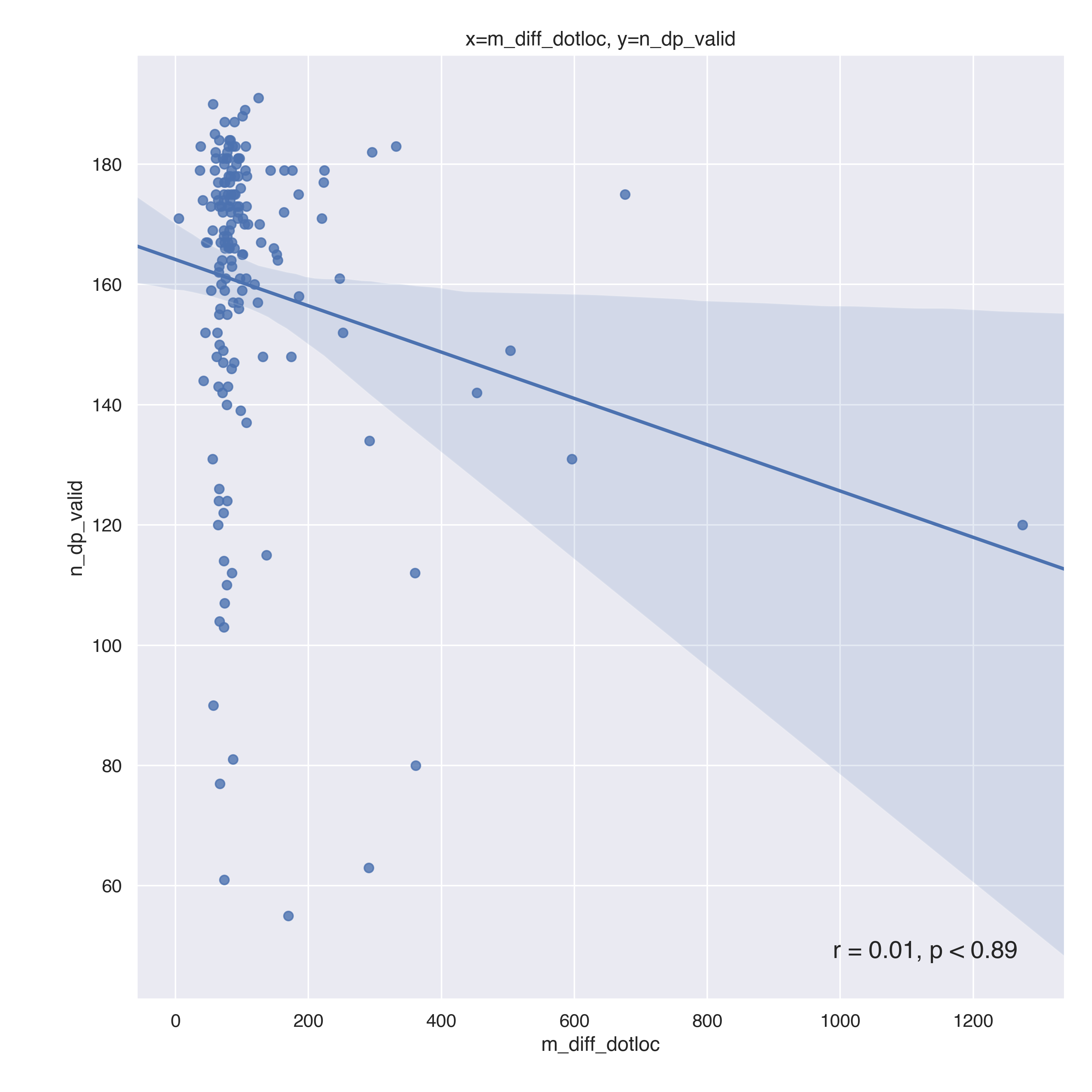
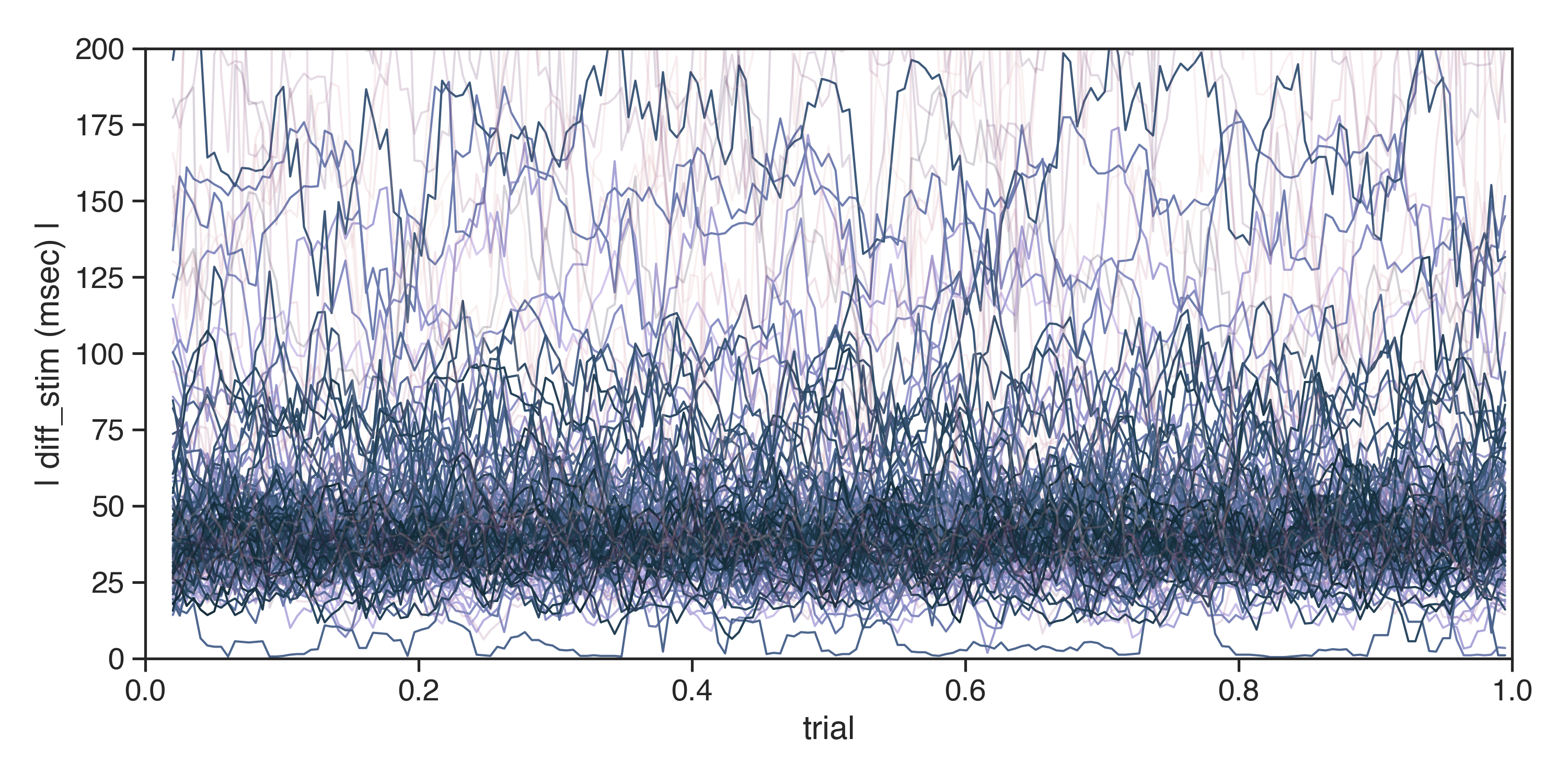
Data collection
md skfsd sd lmfkfds mdskfsd sdl mfkfds mdskfs d sdlmfkfds mdskfsd sdl mfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sd lmfkfds mdskfsd sdlmfkfds mdskfsd sdl mfkfds mdskfsd sdl mfkfds mdskfsd sdlmfkfds mds kfsd sdlmfkfds mdskfsd sd lmfkfds md skfsd sdlmf kfds mdskfsd sd lmfkfds mdskfsd sdlm fkfds mdskfsd sdlm fkfds md skfsd sdl mfkfds mdskfsd sdl mfkfds mdskfsd sdlmfkfds mdskfsd sd lmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskf sd sdl mfkfds mdskfsd sdlmf kfds mds kfsd sdl mfkfds mdskfsd sdlmfkfds.
mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds
mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds
Data processing
mdskfsd sdlmfkfds mds kfsd sdlmfkfds mdskfsd sdl mfkfds mdskf sd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdl mfkfds mds kfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sd lmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlm fkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds.
#run correlation matrix
df_corr = df[['dp_bias','n_dp_valid','var_dp_bias',
'gaze_bias','n_gaze_valid','var_gaze_bias','final_gaze_bias',
'rrs_brooding','cesd_score',
'm_rt','m_diff_dotloc','m_diff_stim',
'luminance']].loc[df['nested'] == 'subject']
#file
file = 'corr_matrix'
method = 'spearman'
title = string.capwords('%s correlation coefficient matrix (p-value).'%(method))
path = path_['output'] + "/analysis/html/%s.html"%(file)
corr_matrix = plot.corr_matrix(config=config, df=df_corr, path=path, title=title, method=method)
del path, corr_matrix, file, title, method
mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds

mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds
mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds mdskfsd sdlmfkfds
Model
| index | term | B | SE | t | df | Pr(>|t|) |
|---|---|---|---|---|---|---|
| 1 | (Intercept) | 6.0775 | 0.1605 | 37.8551 | 139.448161 | 0.0 |
| 2 | osmsos | 0.6379 | 0.2513 | 2.538 | 136.000295 | 0.0123 |
| 3 | trialTypepofa | 0.0138 | 0.0214 | 0.6454 | 27049.127397 | 0.5187 |
| 4 | TrialNum | 0.1126 | 0.0467 | 2.4106 | 136.999935 | 0.0173 |
| index | sigma | logLik | AIC | BIC | REMLcrit | df.residual |
|---|---|---|---|---|---|---|
| 1 | 1.760331 | -54598.524964 | 109213.049928 | 109278.774094 | 109197.049928 | 27316 |
Participants with 'Dotloc' or 'Stimulus' Onset Error median above 3SD (n = [17, 25, 49, 54, 59, 77, 80, 89, 112, 123, 138, 140, 150, 153, 180, 182, 185, 212, 221, 248, 250, 256, 262, 269, 292, 294, 298, 319, 999999, 111111, 156], 18.7%) were excluded from analysis (see methods).
We employed linear mixed effects models with random intercepts and slopes using the lmer() function in the lme4 R package (R Core Team, 2013; Bates, Mächler, Bolker, & Walker, 2015). For our model, Operating System, Stimulus (IAPS, POFA), and Trial. were included as fixed effects. Random effects for Trial, and Participant. were included in the model to account for their respective variation in their slopes and intercepts. Stimulus Onset Error was used as the outcome measure.
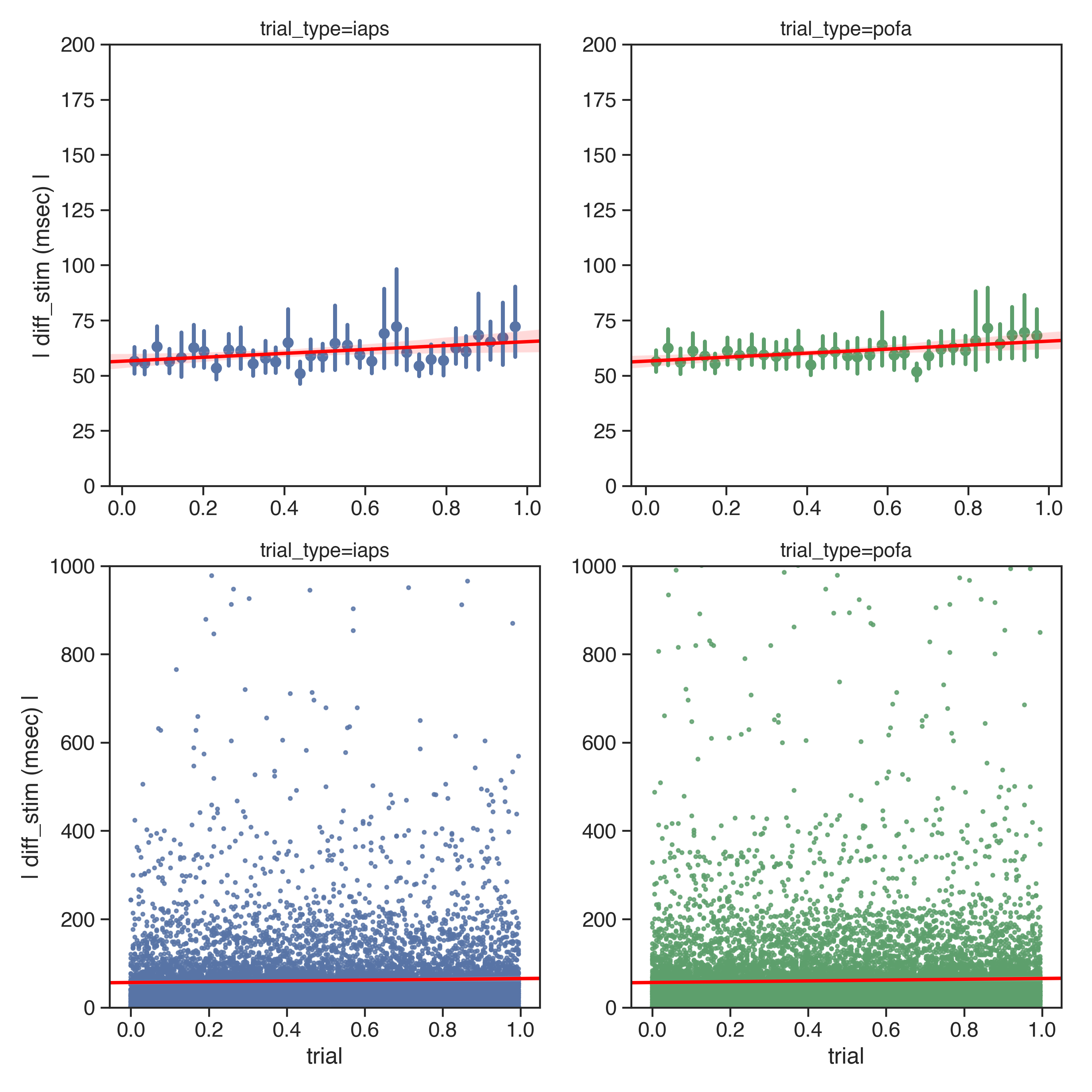
Each individual subjects individual trend is indicated here. Participants with 'Dotloc' or 'Stimulus' onset error rate 3 SD above the median are indicated here with a semi-opaque line. The graph has been clipped at y = 200 for displaying purposes.
Data is either unbinned (c,d) or binned into 33 discrete evenly-sized groups (a,b). The model is still fit using the original data. No participants have been excluded for this analysis. The binned graph has been clipped at y = 1000 for displaying purposes.
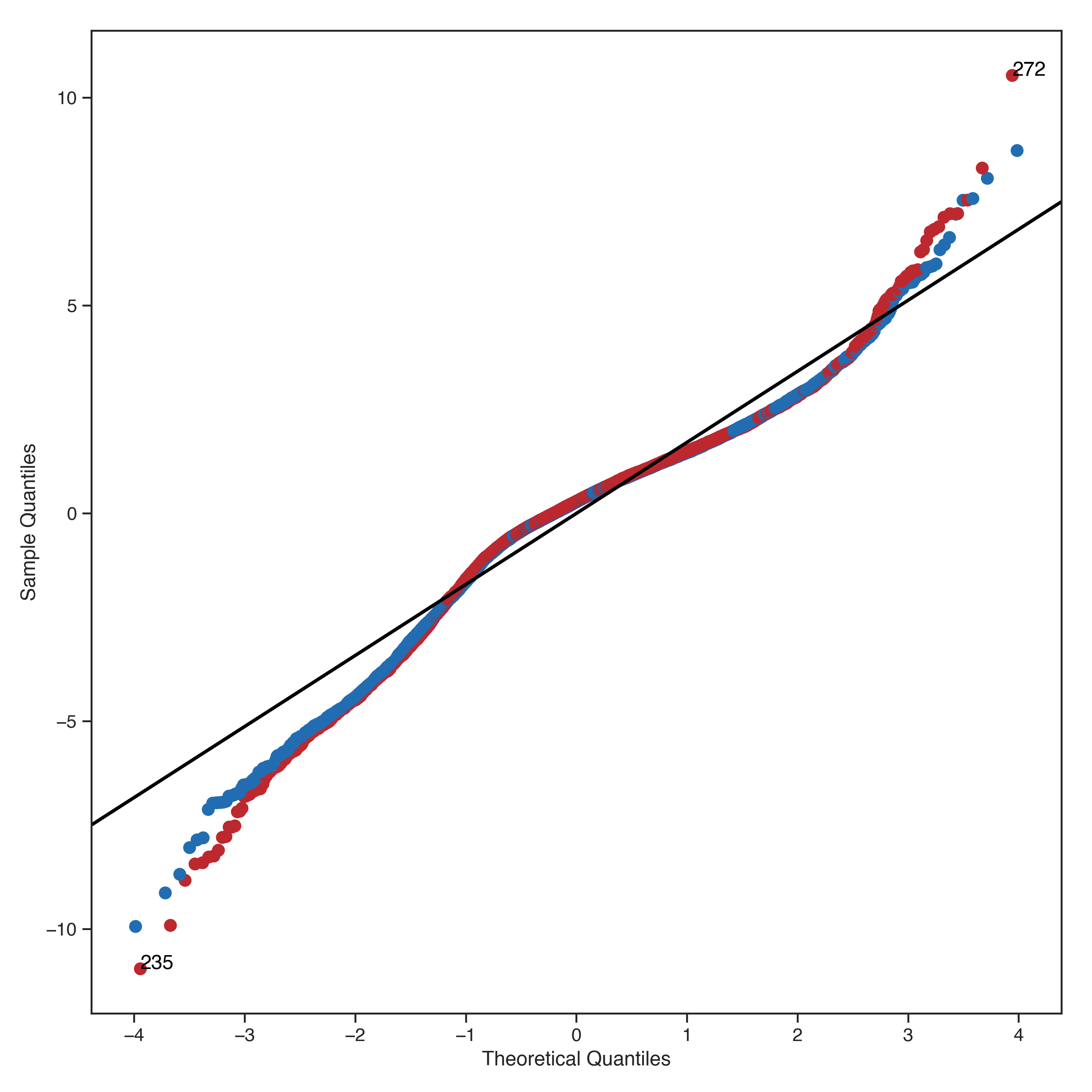
The Normal Q-Q plot compares the standardized residuals against the theoretical quantiles from a standard normal distribution. If the model residuals are normally distributed, then the points on this graph will be plotted in a generally straight line.
effects = {}
#----load data
p = path_['output'] + "/analysis/error.csv"
#df_error = pd.read_csv(p_error, float_precision='high')
df_ = pd.read_csv(p, float_precision='high')
#----parameters
# dependent variable
y = 'diff_stim','diff_dotloc'
# fixed effects
effects['fixed'] = {
'os': 'categorical',
'trialType': 'categorical',
'TrialNum': 'factorial'
}
# random effects
effects['random'] = {
'TrialNum': 'factorial',
'participant': 'factorial',
}
#----save data for access by R and for calculating dwell time
csv = "onset_data.csv"
#----run model
# path
p = path_['output'] + "/analysis/html/model/lmer/"
# formula
f = "sqrt(%s) ~ os + trialType + TrialNum + (1+TrialNum|participant)"%(_y)
# run
lmer_, lmer_result, lmer_r, html = model.lmer(config=config_, df=df_, y=_y, f=f,
exclude=exclude, csv=csv, path=p, effects=effects)
#-----delete
del y, _y, f, csv, p